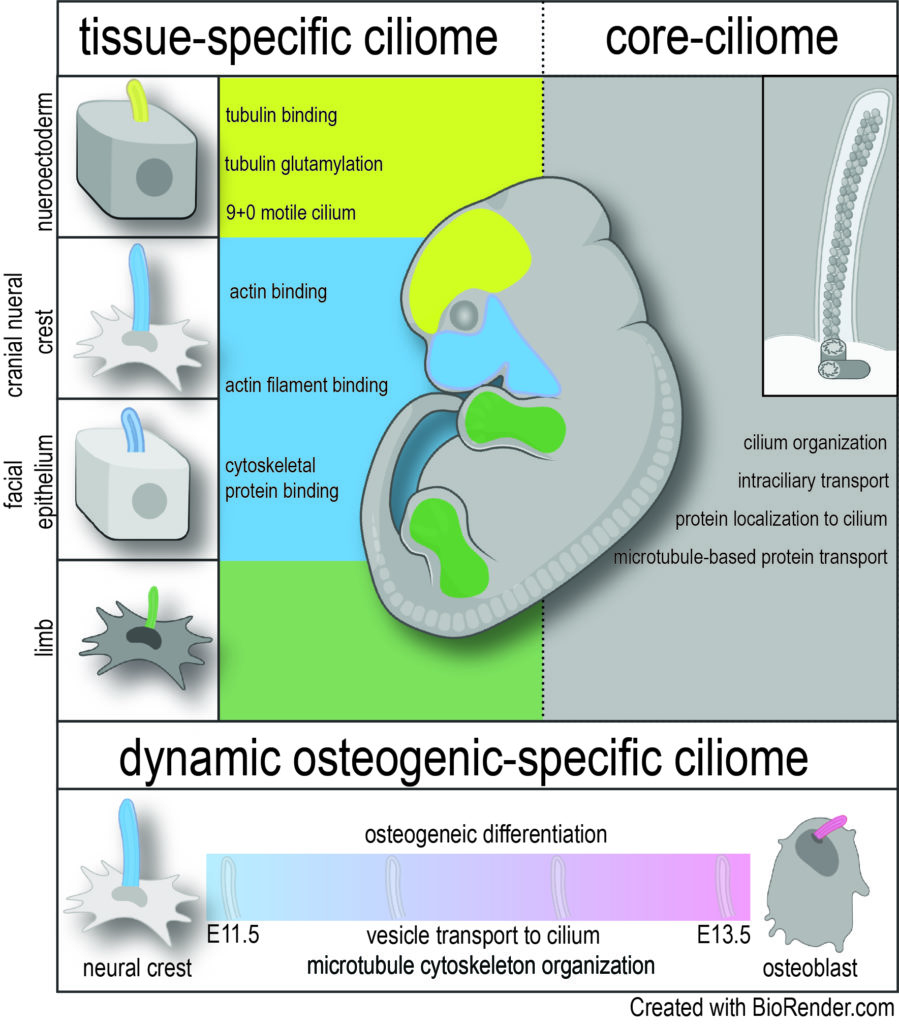
Primary cilia are nearly ubiquitous organelles that transduce molecular and mechanical signals. While the basic structure of the cilium and the cadre of genes that contribute to ciliary formation and function (the ciliome) are believed to be evolutionarily conserved, the presentation of ciliopathies with narrow, tissue-specific phenotypes and specific molecular readouts suggests an unappreciated heterogeneity within this organelle.
Herein, we consolidated three established ciliary databases: The Syscilia Gold Standard, CiliaCarta, and Cildb [1,2] with two ciliogenesis modulator screens [3, 4] to generate a curated ciliome. Expression of genes that make up the ciliome was then profiled across various embryonic tissues and time points to test the hypothesis that the ciliome is heterogenous throughout development.
Here, we provide access to our transcriptomics resources detailing tissue-specific ciliome heterogeneity in an easily searchable database.
- Arnaiz, O., et al., Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia, 2014. 3: p. 9.
- Arnaiz, O., et al., Cildb: a knowledgebase for centrosomes and cilia. Database (Oxford), 2009. 2009: p. bap022.
- Kim, J., et al., Functional genomic screen for modulators of ciliogenesis and cilium length. Nature, 2010. 464(7291): p. 1048-51.
- Wheway, G., et al., An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol, 2015. 17(8): p. 1074-1087.